
|
I am a Staff Research Scientist at Google DeepMind. Previously I was a PhD student at the Bayesian Methods Research Group under the supervision of Dmitry Vetrov. My research interests include deep learning, Bayesian methods, and machine learning for biology. I have worked on AlphaFold which has been recognized as the solution to the protein folding problem. Email / CV / Google Scholar / Twitter / Github |
|
July 2021: We have published a Nature paper describing AlphaFold, as well as the source code and a database of predicted structures. May 2021: I have been promoted to Staff Research Scientist. Nov 2020: We have announced a new version of AlphaFold at CASP14. |
|
|
|
Mihaly Varadi, Stephen Anyango, Mandar Deshpande, Sreenath Nair, Cindy Natassia, Galabina Yordanova, David Yuan, Oana Stroe, Gemma Wood, Agata Laydon, Augustin Žídek, Tim Green, Kathryn Tunyasuvunakool, Stig Petersen, John Jumper, Ellen Clancy, Richard Green, Ankur Vora, Mira Lutfi, Michael Figurnov, Andrew Cowie, Nicole Hobbs, Pushmeet Kohli, Gerard Kleywegt, Ewan Birney, Demis Hassabis, Sameer Velankar Nucleic Acids Research, 2022 paper / AlphaFold Protein Structure Database |
|
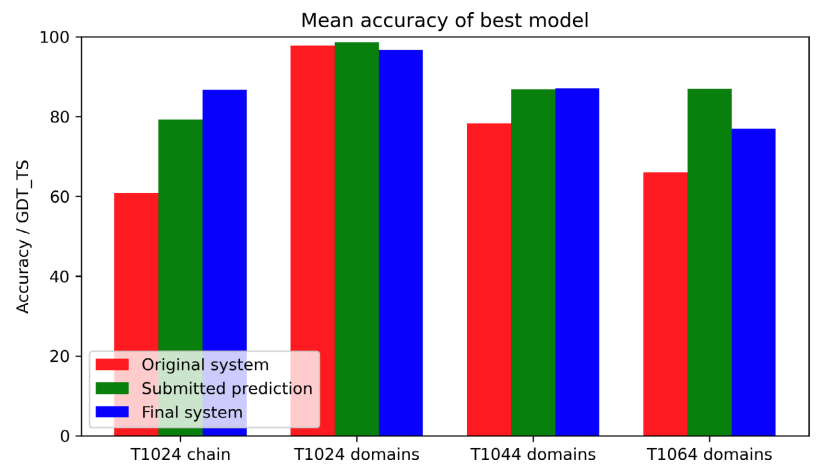
John Jumper*, Richard Evans*, Alexander Pritzel*, Tim Green*, Michael Figurnov*, Olaf Ronneberger*, Kathryn Tunyasuvunakool*, Russ Bates*, Augustin Žídek*, Anna Potapenko*, Alex Bridgland*, Clemens Meyer*, Simon A. A. Kohl*, Andrew J. Ballard*, Andrew Cowie*, Bernardino Romera-Paredes*, Stanislav Nikolov*, Rishub Jain*, Jonas Adler, Trevor Back, Stig Petersen, David Reiman, Ellen Clancy, Michal Zielinski, Martin Steinegger, Michalina Pacholska, Tamas Berghammer, David Silver, Oriol Vinyals, Andrew W Senior, Koray Kavukcuoglu, Pushmeet Kohli, Demis Hassabis* Proteins: Structure, Function, and Bioinformatics, 2021 paper |
|
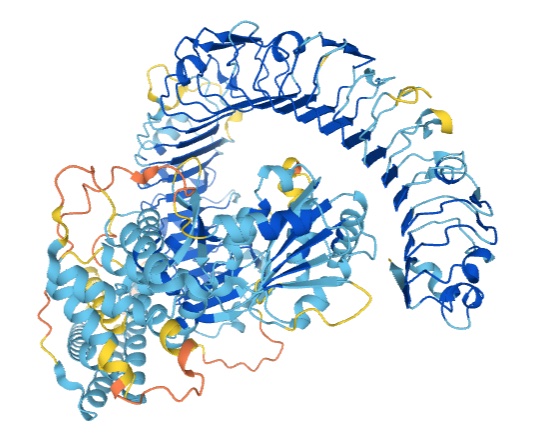
Kathryn Tunyasuvunakool, Jonas Adler, Zachary Wu, Tim Green, Michal Zielinski, Augustin Žídek, Alex Bridgland, Andrew Cowie, Clemens Meyer, Agata Laydon, Sameer Velankar, Gerard J. Kleywegt, Alex Bateman, Richard Evans, Alexander Pritzel, Michael Figurnov, Olaf Ronneberger, Russ Bates, Simon A. A. Kohl, Anna Potapenko, Andrew J. Ballard, Bernardino Romera-Paredes, Stanislav Nikolov, Rishub Jain, Ellen Clancy, David Reiman, Stig Petersen, Andrew W. Senior, Koray Kavukcuoglu, Ewan Birney, Pushmeet Kohli, John Jumper, Demis Hassabis Nature, 2021 paper / blog post: Putting the power of AlphaFold into the world’s hands |
|
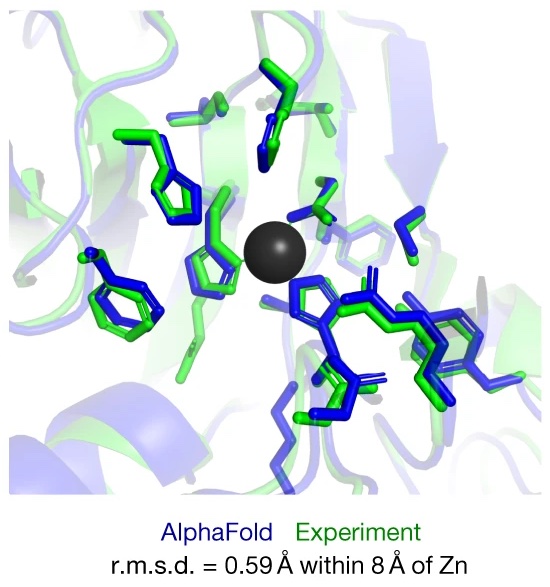
John Jumper*, Richard Evans*, Alexander Pritzel*, Tim Green*, Michael Figurnov*, Olaf Ronneberger*, Kathryn Tunyasuvunakool*, Russ Bates*, Augustin Žídek*, Anna Potapenko*, Alex Bridgland*, Clemens Meyer*, Simon A. A. Kohl*, Andrew J. Ballard*, Andrew Cowie*, Bernardino Romera-Paredes*, Stanislav Nikolov*, Rishub Jain*, Jonas Adler, Trevor Back, Stig Petersen, David Reiman, Ellen Clancy, Michal Zielinski, Martin Steinegger, Michalina Pacholska, Tamas Berghammer, Sebastian Bodenstein, David Silver, Oriol Vinyals, Andrew W. Senior, Koray Kavukcuoglu, Pushmeet Kohli, Demis Hassabis* Nature, 2021 paper / code / case study of AlphaFold / blog post "AlphaFold: a solution to a 50-year-old grand challenge in biology" / blog post "Putting the power of AlphaFold into the world's hands" |
|
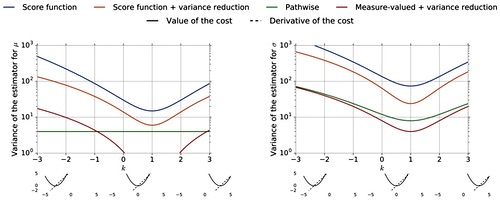
Shakir Mohamed*, Mihaela Rosca*, Michael Figurnov*, Andriy Mnih* JMLR, 2020 arxiv / code (TensorFlow) / code (JAX) |
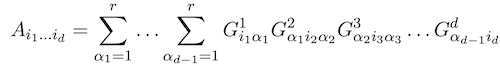
|
Alexander Novikov, Pavel Izmailov, Valentin Khrulkov, Michael Figurnov, Ivan V Oseledets JMLR Open Source Software, 2020 arxiv / code (TensorFlow) / Python package |
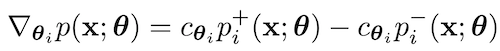
|
Mihaela Rosca*, Michael Figurnov*, Shakir Mohamed, Andriy Mnih Bayesian Deep Learning (NeurIPS Workshop) oral, 2019 paper / talk (11 minutes) / code (TensorFlow) / code (JAX) |

|
Oleg Ivanov, Michael Figurnov, Dmitry Vetrov ICLR, 2019 arxiv / poster / code |
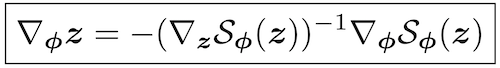
|
Michael Figurnov, Shakir Mohamed, Andriy Mnih NeurIPS spotlight, 2018 arxiv / poster / spotlight video (3 minutes) / spotlight slides / code is integrated into TensorFlow and TensorFlow Probability, eg: Gamma distribution, Beta distribution, Dirichlet distribution, Von Mises distribution, mixture of distributions (set reparameterize=True) |
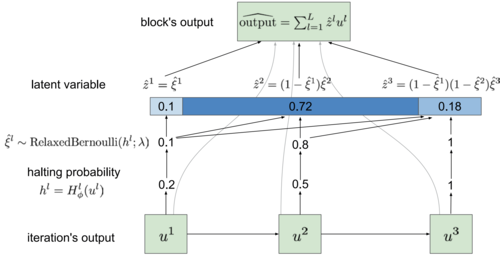
|
Michael Figurnov, Artem Sobolev, Dmitry Vetrov Bulletin of the Polish Academy of Sciences; Deep Learning: Theory and Practice, 2018 paper / arxiv (slightly older version) |
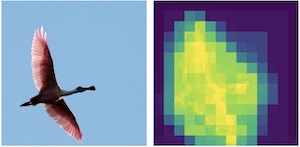
|
Michael Figurnov, Maxwell D. Collins, Yukun Zhu, Li Zhang, Jonathan Huang, Dmitry Vetrov, Ruslan Salakhutdinov CVPR, 2017 arxiv / poster / code (TensorFlow) |
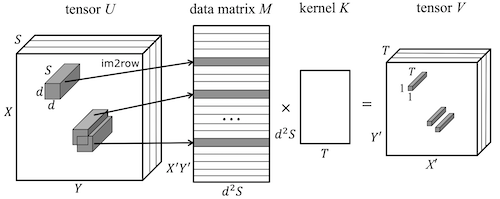
|
Michael Figurnov, Aijan Ibraimova, Dmitry Vetrov, Pushmeet Kohli NeurIPS, 2016 arxiv / poster / code (Caffe) / code (MatConvNet) |
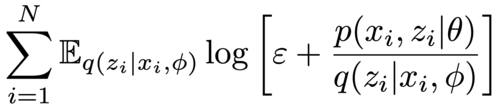
|
Michael Figurnov, Kirill Struminsky, Dmitry Vetrov Advances in Approximate Bayesian Inference, NeurIPS, 2016 arxiv |
|
|

|
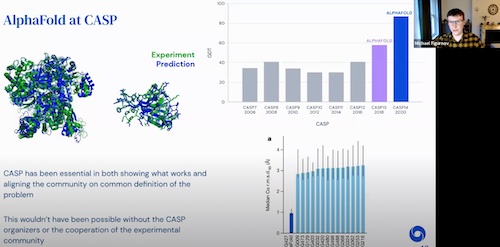
NASSMA 2022 AI4Science, 2022 video |
|
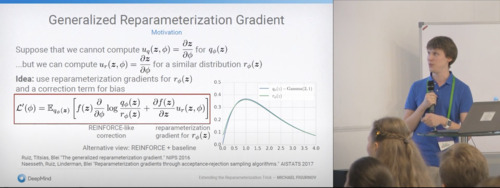
Bayesian Methods Research Group Seminar, 2021 video / slides |
|
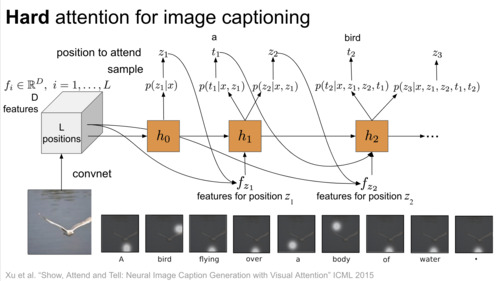
DeepBayes Summer School, 2018 video / slides |
|
DeepBayes Summer School, 2017 video (in Russian) / slides |
|
Thanks to Jon Barron for the template! |